Fungal regulatory systems directing mammalian host colonization
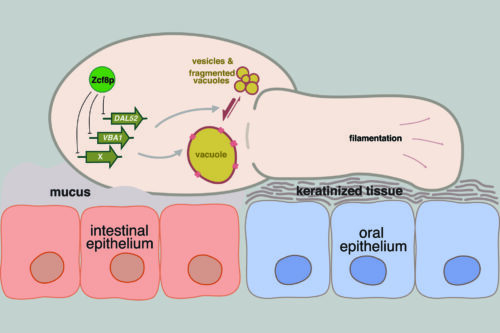
The yeast Candida albicans inhabits the intestine and other mucosal surfaces of humans. To identify gene regulatory circuits that enable this fungus to associate with its host, our lab has screened a library of C. albicans transcriptional regulator deletion strains in mouse models of intestinal and mucosal colonization.
We have identified about a dozen regulators that contribute to the in vivo fitness of the yeast in these niches. We employ genome-wide molecular biology approaches (RNA-Seq and ChIP-Seq) as entry points to explore the function(s) of the transcription regulators identified in our genetic screens. Biochemical, molecular and cell biology experiments are then devised to elucidate the role that each regulator plays in the biology of C. albicans.
Interkingdom microbial interactions in the mammalian gut
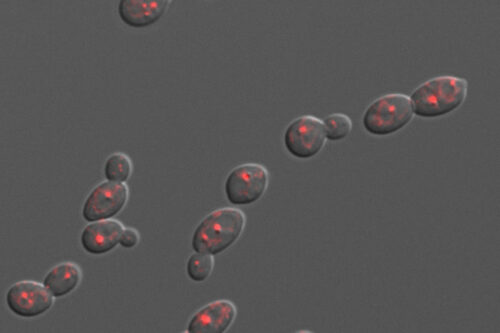
C. albicans causes thousands of disseminated, life-threatening infections in the United States every year. Such infections are preceded by an expansion of the fungus in the patients’ gut and by microbial dysbiosis. To investigate how co-inhabiting microbes may shape C. albicans proliferation in the gastrointestinal tract, our lab has established a gnotobiotic mouse model of C. albicans gut colonization.
In this experimental system, the fungus is gavaged in mice that have been raised germ free. Gut commensal bacteria, either alone or as part of defined communities, are also introduced. We use a combination of imaging tools and genetic screens to define how bacteria influence C. albicans proliferation in the mammalian intestine.
Responses elicited by commensal fungi in the mouse intestine
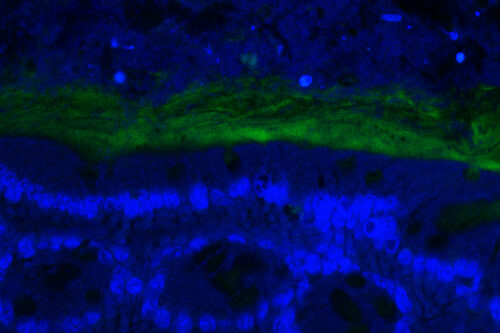
Disbalances between host and gut commensal fungi have also been linked to inflammatory bowel diseases, hinting at the occurrence of meaningful signaling between gastrointestinal tract and its fungal dwellers. Yet the mechanisms whereby commensal fungi are sensed in the mammalian intestine remain largely undefined.
We are employing the gnotobiotic mouse model described above as a tool to interrogate the responses specifically elicited by the commensal fungus C. albicans while residing in the gastrointestinal tract.

